Data Processing Assistant for Resting-State fMRI (DPARSF) V2.3
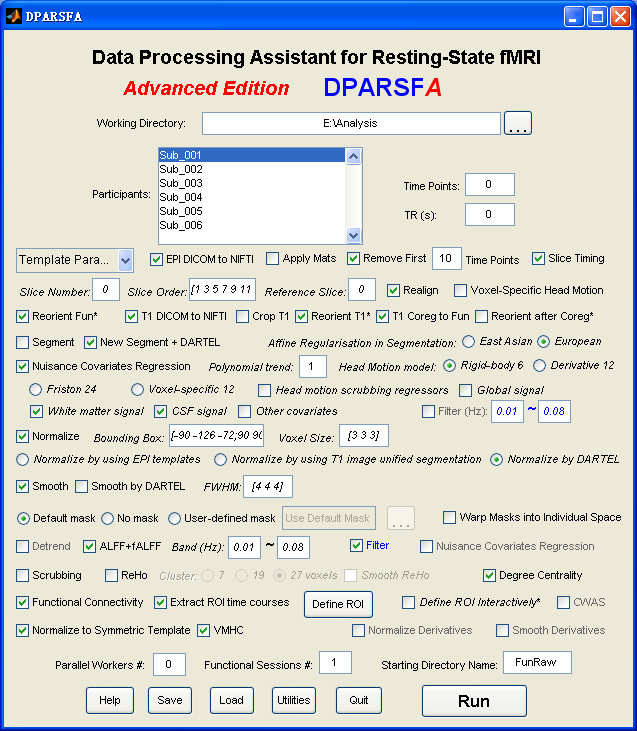 |
Data Processing Assistant for Resting-State fMRI (DPARSF) is a convenient plug-in software based on SPM and REST. You just need to arrange your DICOM files, and click a few buttons to set parameters, DPARSF will then give all the preprocessed (slice timing, realign, normalize, smooth) data, functional connectivity, ReHo, ALFF/fALFF, degree centrality, voxel-mirrored homotopic connectivity (VMHC) results. DPARSF can also create a report for excluding subjects with excessive head motion and generate a set of pictures for easily checking the effect of normalization. You can use DPARSF to extract ROI time courses efficiently if you want to perform small-world analysis. DPARSF basic edition is very easy to use while DPARSF advanced edition (alias: DPARSFA) is much more flexible and powerful. DPARSFA can parallel the computation for each subject, and can be used to reorient your images interactively or define regions of interest interactively. You can skip or combine the processing steps in DPARSF advanced edition freely. Please download a MULTIMEDIA COURSEto know more about how to use this software. Add DPARSF's directory to MATLAB's path and enter "DPARSF" or "DPARSFA" in the command window to enjoy DPARSF basic edition or advanced edition. The latest release is DPARSF_V2.3_130615. |
New features of DPARSF_V2.3_130615:
1. Apply downloaded reorient matrices. Given the amount of time and effort for interactive reorienting, the reorient matrices for online data such as 1000 Functional Connectomes Project (FCP) (Biswal et al., 2010) and Autism Brain Imaging Data Exchange (ABIDE) (Di Martino et al., 2013) could be downloaded and applied automatically. Please find here for “DownloadedReorientMats”, and then tick the checkbox "Apply Mats".
2. Save information of TR, Slice Number, Time Points and Voxel Size into TRInfo.tsv (under working directory) file for checking data correctness.
3. The slice order type could be specified for each participant into SliceOrderInfo.tsv (under working directory) file, thus allow different slice timing correction for different participants in a batch mode. Please find instructions for setting SliceOrderInfo.tsv from {DPARSF}/Docs/SliceOrderInfo.tsv_Instruction.txt.
4. The output format of DICOM to NIfTI were changed to 4D .nii images.
5. The midline of VMHC results were set to zero.
Biswal, B.B., Mennes, M., Zuo, X.N., Gohel, S., Kelly, C., Smith, S.M., Beckmann, C.F., Adelstein, J.S., Buckner, R.L., Colcombe, S., Dogonowski, A.M., Ernst, M., Fair, D., Hampson, M., Hoptman, M.J., Hyde, J.S., Kiviniemi, V.J., Kotter, R., Li, S.J., Lin, C.P., Lowe, M.J., Mackay, C., Madden, D.J., Madsen, K.H., Margulies, D.S., Mayberg, H.S., McMahon, K., Monk, C.S., Mostofsky, S.H., Nagel, B.J., Pekar, J.J., Peltier, S.J., Petersen, S.E., Riedl, V., Rombouts, S.A., Rypma, B., Schlaggar, B.L., Schmidt, S., Seidler, R.D., G, J.S., Sorg, C., Teng, G.J., Veijola, J., Villringer, A., Walter, M., Wang, L., Weng, X.C., Whitfield-Gabrieli, S., Williamson, P., Windischberger, C., Zang, Y.F., Zhang, H.Y., Castellanos, F.X., Milham, M.P., 2010. Toward discovery science of human brain function. Proc Natl Acad Sci U S A 107, 4734-4739.



byz
Thu, 10/31/2013 - 02:40
Permalink
DPARSFA怎样导入数据
严老师:
您好!我最近在学习DPARSFA,但一开始就遇到了问题,我将数据文件夹FunImg的上一级文件夹作为数据路径放入MATLAB,但是在DPARSFA中无法导入数据,DPARSFA和DPARSF放数据的路径Working Directory设置不一样吗?DPARSFA应该怎么设置Working Directory呢?
望老师能给与指导,谢谢老师!
YAN Chao-Gan
Thu, 10/31/2013 - 02:52
Permalink
Hi,
Hi,
You'll see the subjects after you setting up "Starting Directory Name".
Best,
Chao-Gan
Philip.xu
Thu, 10/31/2013 - 11:20
Permalink
回复: [RFMRI] Data Processing
byz
Fri, 11/01/2013 - 01:59
Permalink
已经解决了,谢谢您,以后我会仔细看视频的。
已经解决了,谢谢您,以后我会仔细看视频的。
wisepig119
Wed, 11/06/2013 - 01:26
Permalink
DPARSF报错
YAN Chao-Gan
Wed, 11/06/2013 - 16:00
Permalink
Re: [RFMRI] Data Processing
wisepig119
Sat, 11/09/2013 - 02:49
Permalink
这样啊,我从预处理开始就开着电脑,然后就睡觉去了
这样啊,我从预处理开始就开着电脑,然后就睡觉去了,早上起来就发现这样了,不过我后来接着重新运算了ALFF,就没有什么问题了,谢谢超赣兄!
martin.dyrba
Sun, 11/24/2013 - 14:54
Permalink
ROI_OrderKey bugfix
Dear Chao-Gan,
I noticed a problem that the lastly defined ROI and it's ROI indices get lost when writing the ROI_OrderKey_*.csv files for the 'extract ROI signal' step. Could you please remove the following line at the bottom part of y_ExtractROISignal.m ?
MaskROILabel{1,length(ROIDef)} = []; % Force the undefined cells to emptyRegards,
Martin
YAN Chao-Gan
Mon, 11/25/2013 - 04:47
Permalink
Hi Martin,
Hi Martin,
Thanks a lot for your report!
I fixed it by changing into:
if size(MaskROILabel,2) < length(ROIDef) %YAN Chao-Gan, 131124. To avoid if the labels of the last ROI has been defined.
MaskROILabel{1,length(ROIDef)} = []; % Force the undefined cells to empty
end
This will be updated in the next version.
Thanks,
Chao-Gan
hmontazery
Mon, 04/21/2014 - 05:25
Permalink
How read the rest fmri data of ADHD-200 with nii suffix in DPARS
Dear Mrs/Mr
Please, if possible, guide me how I can read the rest fmri data of ADHD-200 with nii suffix in DPARSF?
In your matlab based DPARSF, I can't load the my NIFTI file. Do I get the processed ADHD-200 competition data? I need the time course or BOLD signal for my research.
I will be grateful if you could help me.
Best regards,
juefirst
Mon, 04/21/2014 - 06:12
Permalink
Dear Hussain Montazery Kordy,
Dear Hussain Montazery Kordy,
I've met this problem recently and solved already. I post the solution here, hope to help you.
ADHD-200 data are 4-D, so you have to covert it to 3-D first.
funcdir='/public/home/FunImg';
cd ([funcdir '/']);
dirlist=dir(cd);
for j=3:length(dirlist)
restdir=[funcdir,filesep,dirlist(j).name,filesep];
cd (restdir);
PI='rest.nii';
PI=PI(1:end-3);
PO='.nii';
[Data, vox,Head] = rest_readfile(PI,'all');
if size(Data, 4)>1
for i=1:size(Data,4)
Index=['000',num2str(i)];
Index=Index(end-3:end);
[Path, fileN, extn] = fileparts(PO);
POout=[Path,filesep,fileN,'_',Index,extn];
rest_WriteNiftiImage(Data(:,:,:,i),Head,POout(end-7:end));
end
end
tycaowei
Mon, 07/14/2014 - 02:31
Permalink
dparsf
严老师,您好
我在使用dparsf处理数据,我先处理了一个人的数据是可以的,但是批处理时在normalize后老是报index ceceeds matrix dimensions的错误,请指教!
谢谢!
YAN Chao-Gan
Mon, 07/14/2014 - 02:33
Permalink
Hi, What's the parameter
Hi, What's the parameter setting and error message?
具体的参数设置和报错信息是什么呢?
超赣
mohitindelhi
Mon, 07/14/2014 - 05:30
Permalink
Re: [RFMRI] Data Processing
& Regards
All India Institute of Medical Sciences,
Ansari Nagar, New Delhi-110029
+919968569874
YAN Chao-Gan
Mon, 07/14/2014 - 14:03
Permalink
Hi Ansari,
Hi Ansari,
You can first select the template parameter of "VBM".
Then if you started with NIfTI images instead of DICOM images, change the starting directory name to the folder of "T1Img".
Best,
Chao-Gan
tycaowei
Mon, 07/14/2014 - 08:10
Permalink
回复: [RFMRI] Data Processing
sharky
Fri, 08/15/2014 - 12:24
Permalink
DPARSFA 2.3: Several fMRI sessions and one T1
comment deleted
Gaizhi Li
Tue, 12/08/2015 - 00:41
Permalink
??? Error using ==> y_ExtractROISignal at 188
Gaizhi Li
Tue, 12/08/2015 - 01:00
Permalink
FW: [RFMRI] Data Processing
Sent: Monday, December 07, 2015 7:41 PM
To: rfmri.org@rnet.co
Subject: Re: [RFMRI] Data Processing Assistant for Resting-State fMRI (DPARSF) V2.3
Loose
Wed, 05/25/2016 - 11:49
Permalink
Correlation Analysis with text variable
Hi,
I want to do a correlation analysis between FC-Maps of a ROI and a text variable (duration of illness). What I did so far:
I use DPARSF --> statistical analysis --> Correlation analysis: Add FC-images of the patient-group (which are saved in the folder "FC-FunImgRWSDF) and add a text file with the respective duration of illness (in one column; I have 61 subjects, so 61 lines). The calculation works and I can open the result-file.
The first thing I am wondering about is that when I add the group images (so the folder "FC-FunImgRWSDF") the program immediatly reads in the standardized zFCmaps as well as the FCmaps. However I guess that I only want the zFCmaps to be included in the calculation, right? Do I just have to put the zFCmaps in a seperate folder and then add this folder as the image-folder or is there another way (like in one-sample-t-test, where I just type in a 0 for standardized and a 1 for unstandardized data).
Second, when I look at the result-image with a threshold of p=0.05 I see all significant (but uncorrected) correlations of the FCmap and the text-variable, am I right? If i now want to find out about the exact correlation of a respective cluster in this image, I can use the cluster-report. However this will only show me the correlation of the peak-voxel. Is there a way that the programm can tell me the overall correlation for the respective significant cluster?
YAN Chao-Gan
Fri, 05/27/2016 - 04:28
Permalink
Hi,
Hi,
1. "put the zFCmaps in a seperate folder and then add this folder as the image-folder"
2. If you want the mean correlation of the cluster. You can DPABI->Cluster->Save a single cluster to save an ROI file, and then use DPABI->Utilities->ROI Signals Extractor to extract it.
Best,
Chao-Gan
Loose
Sun, 05/29/2016 - 13:29
Permalink
FC and zFCmaps
Thank you very much for the response. Another question: When I do a one-sample or two-sample t-test, do I always have to put the zFC maps in an extra folder? I thought that in a one-sample-t-test by setting the base to 0 (instead of 1) the program would automatically use just the zFC-maps.
YAN Chao-Gan
Mon, 05/30/2016 - 07:20
Permalink
Re: [RFMRI] Data Processing
Best,
Loose
Wed, 06/01/2016 - 11:48
Permalink
individual zFCmaps
Thanks again :-)
I have another problem: I want to take a look at the individual zFCmaps. It works for almost every subject. However for 5 subjects I get the following error message once I try to open the zFCmaps in DPABI-Viewer:
YAN Chao-Gan
Thu, 06/02/2016 - 06:19
Permalink
This is pretty weird. You can
This is pretty weird. You can upload the images to have a look.
Loose
Wed, 06/08/2016 - 13:55
Permalink
FDR-Correction
Ok, the problem with viewing the single subject zFCmaps didn't occur again :-)
However I have another problem. I did a two-sample-t-Test of zFCmaps and now want to do an FDR-correction. I first opened DPABI-viewer, loaded up the T2-image and then just click in "FDR" and set the q-value to 0.05. Am I right?
I did that and unfortunately no differences were found. However a colleaque of mine is using the same data but a different programm and somehow he finds a group difference after FDR-correction. So I was wondering whether I might do it wrong in DPARSF. Also, the other programm filters the z-FCmaps on a threshold of 0.4 before doing any calculations such as t-tests. Only those voxels which are higher than the threshold of 0.4 will be included in the t-test, is that possible in DPARSF as well?
Greetings
Markus
Loose
Wed, 06/08/2016 - 14:30
Permalink
Additional question
Another question: What threshold for the T-Value would you recommend when looking at the seed-network of one group (using one-sample-test of zFCmaps). For the Default-Mode-Netork for instance and a group-size of N=60 I use a T-value of 8 and the network looks quite good, however I am wondering whether this is the proper threshold for checking network quality.
thanks in advance :-)
YAN Chao-Gan
Mon, 06/13/2016 - 09:02
Permalink
Hi Markus,
Hi Markus,
You can perform one sample t-tests for each group, then combine the significant voxels across the two groups as a mask. This is achievable in DPABI.
For threshold in one sample t-test, you can perform GRF correction. Or some people set at p<1E-5 to get better visualization.
Best,
Chao-Gan
Loose
Mon, 06/13/2016 - 10:23
Permalink
Hi Chao-Gan,
Hi Chao-Gan,
thanks for the reponse :-)
It sounds very interesting, however, I am not sure whether I am doing it right. How do I create a mask upon the significant voxels of both one-sample-t-tests? I tried uploaing both t-test images (of patients and controls) in the DPABI-Viewer, then I set the threshold to 0.000001 and then clicked on save all clusters. This will produce a mask which I then uploaded as a mask when doing the FDR-Correction. Is that the procedure you were talking about or should I do it differently?
Also, which parameters should I use in a GRF-correction. Would you recommend both p=0.05 in Voxel and cluster p-value?
Greetings from Germany
Markus
YAN Chao-Gan
Mon, 06/13/2016 - 23:00
Permalink
Re: [RFMRI] Data Processing
Loose
Tue, 06/14/2016 - 17:09
Permalink
Allright, thanks :-)
Allright, thanks :-)
When I load up the images in the image calculator, do I have to add anything under "Group images"?
I then do FDR correction with the two-sample-t-test and load up the mask in the FDR-correction-window, is that right?
Also, when doing GRF correction, what p-value for cluster respectively voxel would you recommend when using it in a one-sample-t-test to check for network quality. Do I have to load up a mask there as well?
Best,
Markus
YAN Chao-Gan
Tue, 06/14/2016 - 23:00
Permalink
Re: [RFMRI] Data Processing
Loose
Wed, 06/15/2016 - 09:02
Permalink
Additional FDR?
Ok, thanks that was helpful :-)
When I view the one-sample-t-test for a network of one group and set the threshold to p<0.000001 should I do an additional FDR correction or is that unnecessary since the p-value is that small?
Also, when I do an GRF-correction on a one-sample-t-test to check network quality, do I have to upload a mask as well?
YAN Chao-Gan
Tue, 06/21/2016 - 14:27
Permalink
1. I think that's OK without
1. I think that's OK without further FDR correction.
2. Yes, you need to apply a mask.
Best,
Chao-Gan
ayo
Sun, 05/27/2018 - 06:22
Permalink
老師
老師
我使用matlab 2016, spm8, DPARSF_V2.3_130615 run前置分析
但是一直都在FunImgAR此步驟出錯,出錯畫面如下
YAN Chao-Gan
Wed, 05/30/2018 - 06:08
Permalink
Please try the newest version
Please try the newest version of DPABI/DPARSF.
沐溱
Fri, 05/13/2022 - 07:12
Permalink
预处理顺序
Powered by Dr. Yan @ My Research Network