DPABI/DPABISurf/DPARSF Error Handling
DPABI/DPABISurf/DPARSF Error Handling
(中文版在后半部分)
We have noticed that the biggest problem that troubles beginners of functional magnetic resonance imaging is error reporting (Figure 1). Among all the errors, the computer environment setting (software installation) accounted for 40%, and the data sorting problem accounted for 50%. So please be sure to read this guide carefully, it will help you avoid 90% of possible errors, and save your precious time to focus on the analysis of results!
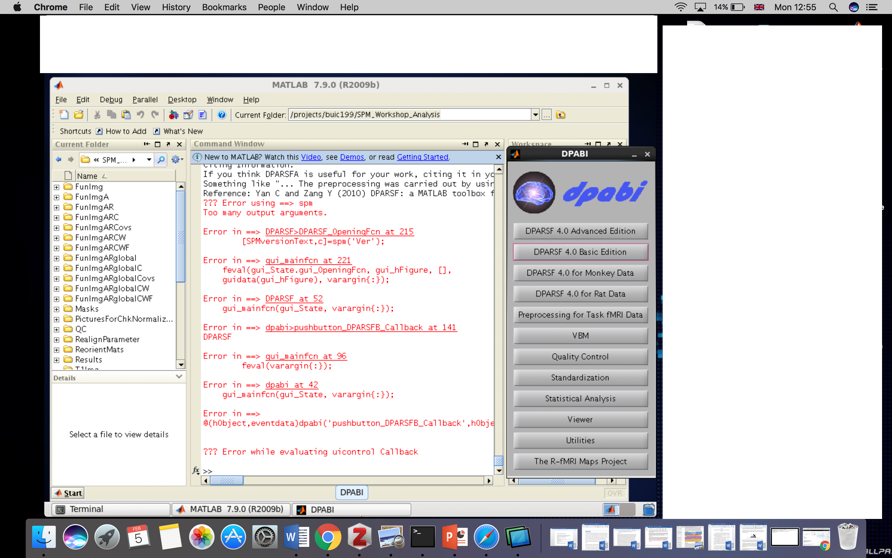
Figure 1. You definitely never want to see this error (“the red”) again
1. Installation of DPABI/DPABISurf
The prerequisites for DPABI/DPABISurf to run smoothly:
(0) A windows/mac/linux computer with relatively decent performance
The equivalent or higher performance of processors than the 7th Gen Intel-i5. More than 16G of the memory. Better to reserve more than 300G disk space.
(1) MATLAB
Many users would like to put SPM or DPABI into the toolbox of MATLAB, then there shouldn’t be any spaces, Chinese characters or special characters in the installation path! Such as
The installation path like "C:\Program files\MATLAB" is absolutely not allowed, as there is a space between Program and files! In addition, if your account name of the operating system ("Administrator" by default) contains Chinese characters, it is recommended to create a new English account (without spaces), and install MATLAB under this account.
(2) SPM
SPM is a free and open source software, please go to http://www.fil.ion.ucl.ac.uk/spm/software/download/ and fill in a table with some simple personal information to download. It is recommended to download SPM12. Remember that the software path should not include any spaces, Chinese characters or special characters!
(3) FSL (please ignore this if you are windows users)
If you are using a computer with a mac or linux system, you need to additionally install another MRI data processing software, FSL, please go to https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FslInstallation, and follow the steps on the web page to download and install it. Alternatively, you can install the docker version of DPABISurf to skip this step.
(4) DPABI/DPARSF
Please go to http://rfmri.org/DPABI to download the latest version of DPABI (including DPARSF).
Installation of SPM and DPABI/DPARSF. Remember that the software path should not include any spaces, Chinese characters or special characters!
After downloading, open MATLAB and click "Set Path" (Figure 2).
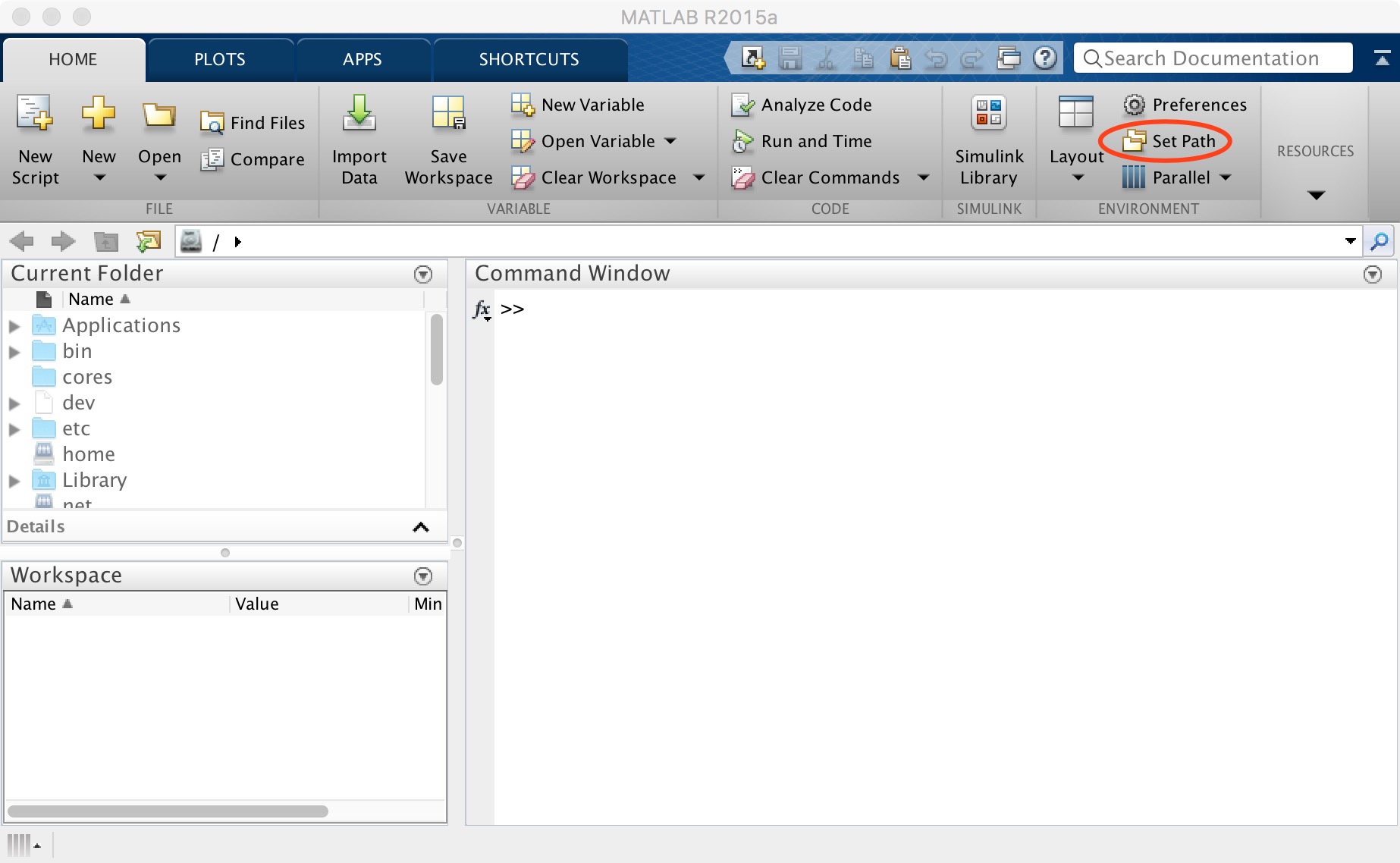
Figure 2. Set path
First, click "default" to restore the path. Next click “Add Folder”to add SPM path (don't "Add with subfolders" for SPM). And then click "Add with subfolders" (Figure 3), find the location where DPABI is stored in the pop-up window and click "open" to add it. Finally, click "save" and close this window.
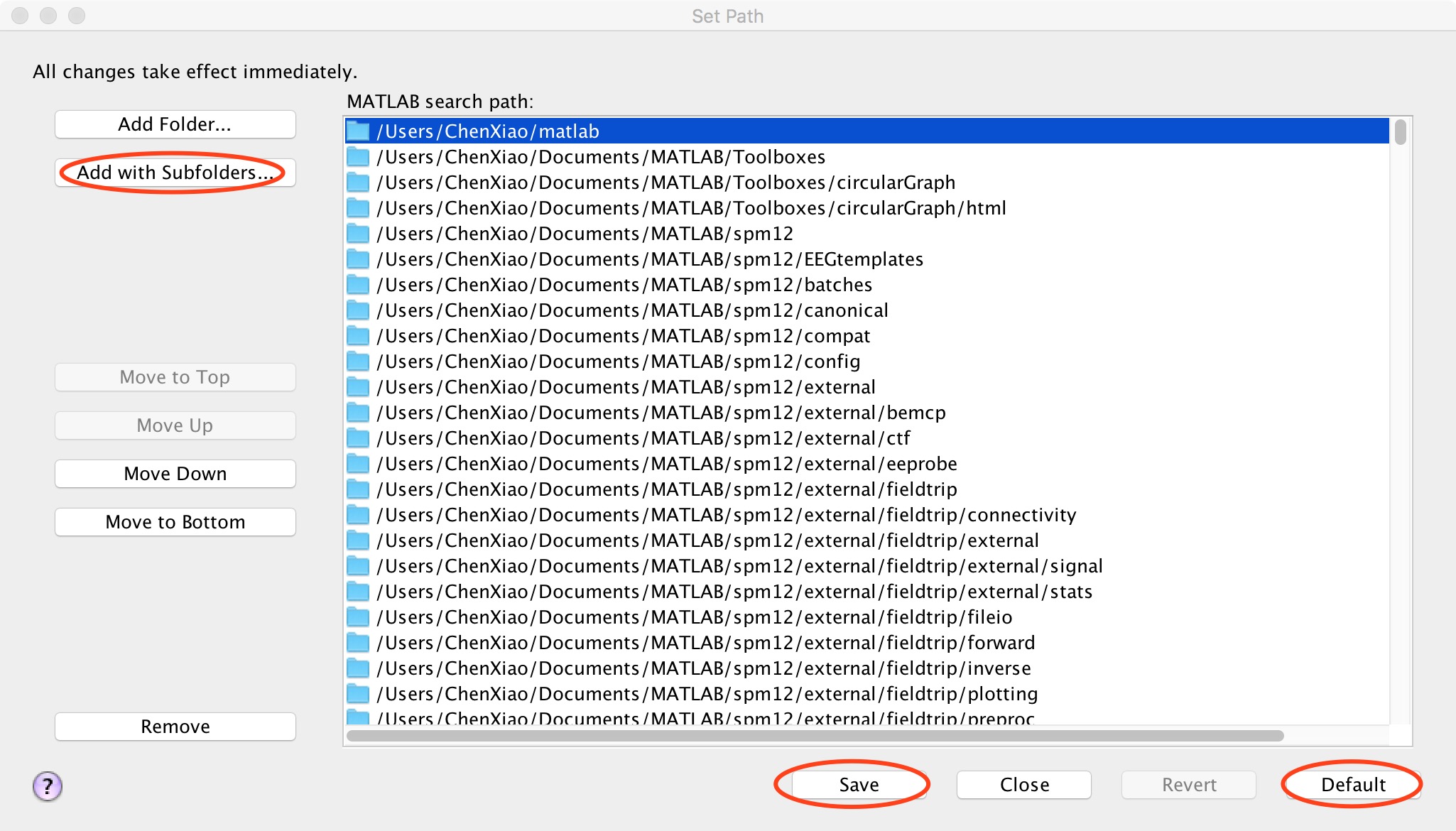
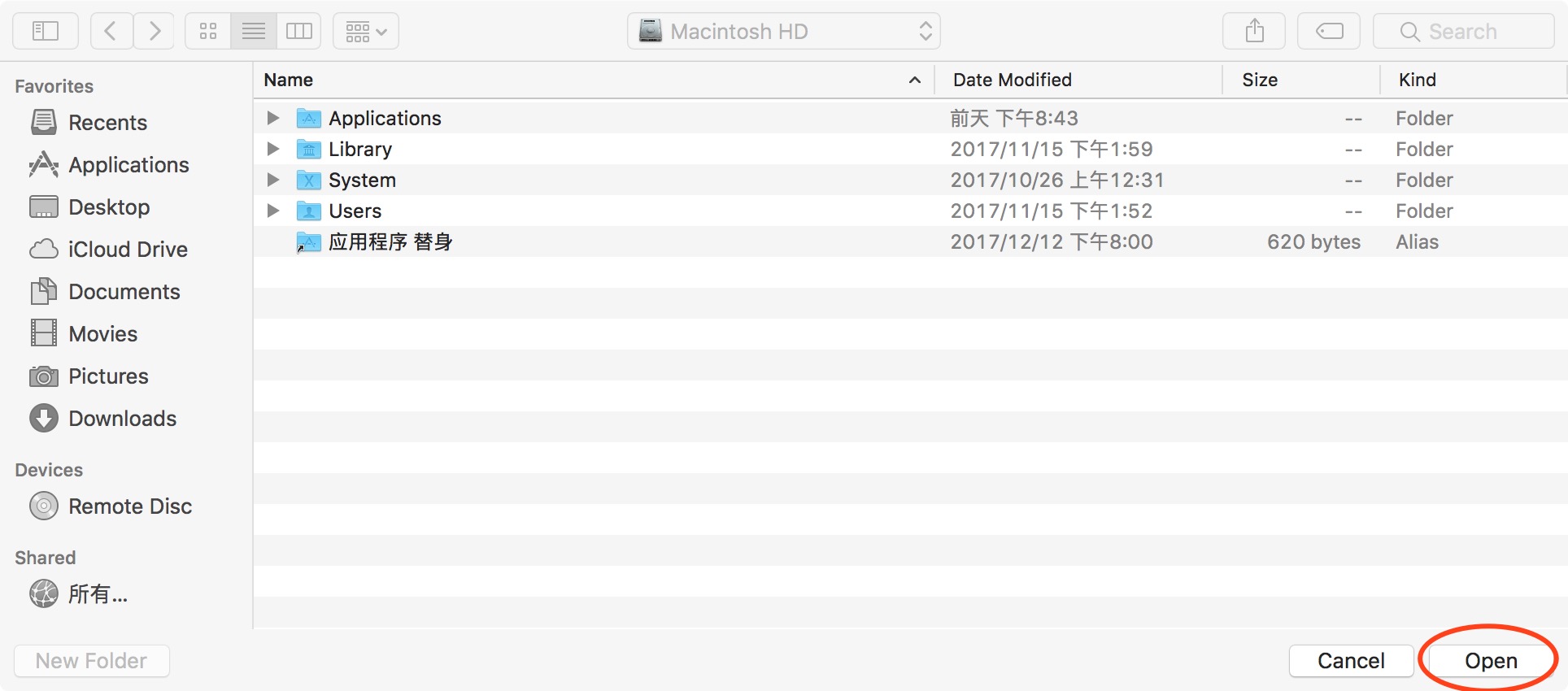
Figure 3. Add with subfolders
After that, you can input "dpabi" in the "command window" and press Enter. When you see the interface as shown below, you successfully completed the installation of DPABI/DPARSF.
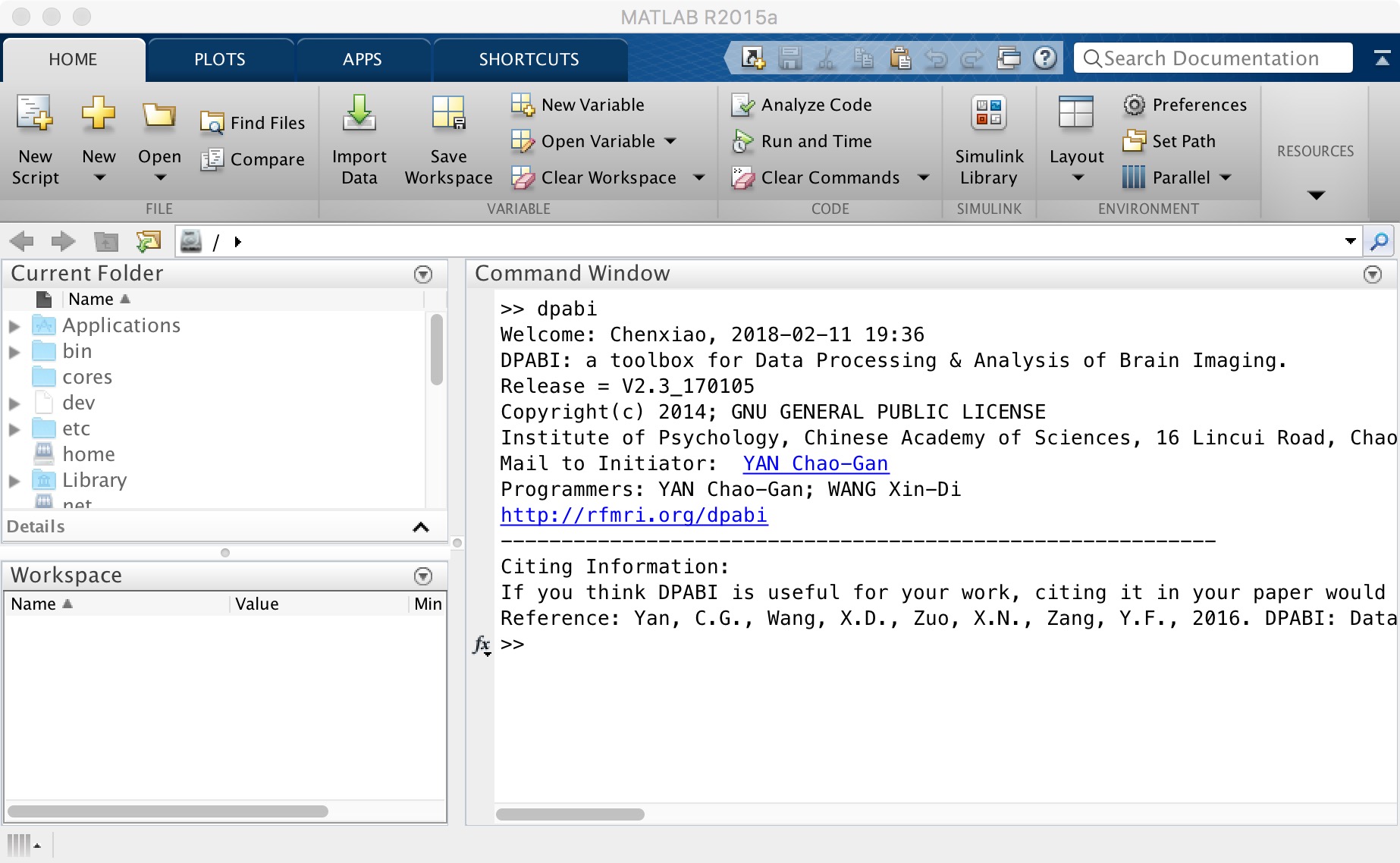
Figure 4. DPABI
2. Data collation
DPABI/DPABISurf is a scientific and efficient R-fMRI pre-processing pipeline. But if you want DPABI/DPARSF to run smoothly, you need to organize your data in strict accordance with the requirements of DPABI/DPARSF. Tips: You can use DPABI->Utilities->DPABI Input Preparer to organize your data! When you are organizing data, please use DPABI->Utilities->Check Data Organization repeatedly to check whether your data organization is qualified! Almost all the errors as "index exceeds dimensions" can be solved by the Check Data Organization!
(1) Create a "working directory"
Create a folder of any name (but it cannot contain any spaces, Chinese characters or special characters!) anywhere you want, which is the "working directory" of preprocessing. Most of the preprocessing operations are under "working directory". This is a very important concept, so please keep it in mind. For demonstration, we created a folder named "dpabi" as the working directory (Figure 5).
Figure 5. Working directory
(2) Copy the MRI data of subjects
Next, we need to the MRI data of subjects under the working directory according to the requirements of DPABI/DPARSF. The following is an example of three subjects:
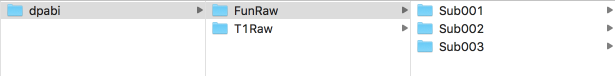
Figure 6. An example of 3 subjects
This figure shows the form of the created folder. The working directory should include folders named "FunRaw" and "T1Raw". Each of them contains three folders: "Sub001", "Sub002", " "Sub003". This example is of the situation of three subjects, please follow this pattern for more subjects.
The “FunRaw” folder contains the functional image data, while the “T1Raw” folder contains the structural images. You need to select the folder containing functional images or structural images from the raw data, and then copy the image data into the corresponding folder.
Note 1: DPABI/DPARSF has very strict requirements on sorting the data format under the working directory. The names of the two folders "FunRaw" and "T1Raw" must be in full compliance with this form, and should not be changed, otherwise the “error” will be reported. Although the name of subject folders such as "Sub001", "Sub002", and "Sub003" are more arbitrary and you can change them according to your needs, you should still keep the names of corresponding subject folders in "FunRaw" and "T1Raw" are exactly the same. Any difference will cause an error. An example is when a student accidentally entered an extra space at the end when naming a folder, which resulted in an error.
Note 2: The example here fits when the raw data you had is in dicom format. If your data is in NIFTI format, please rename the two folders "FunRaw" and "T1Raw" as "FunImg" and "T1Img". The way to judge the dicom/NIFTI format: the number of data files in the dicom format is very large (hundreds to thousands); while the number of data files in the NIFTI format is one for each subject and each sequence is only one (4D .nii file) or hundreds (3D .img/.hdr files).
Note 3: Generally speaking, the raw data is organized according to the sequence and the data of each scan sequence has its corresponding folder. But sometimes the data is completely messy, then you need to use the "Dicom Sorter" function of DPABI to organize the messy raw data into a form that we can understand.
To open Dicom Sorter, please call DPABI and click Utilities->DICOM Sorter
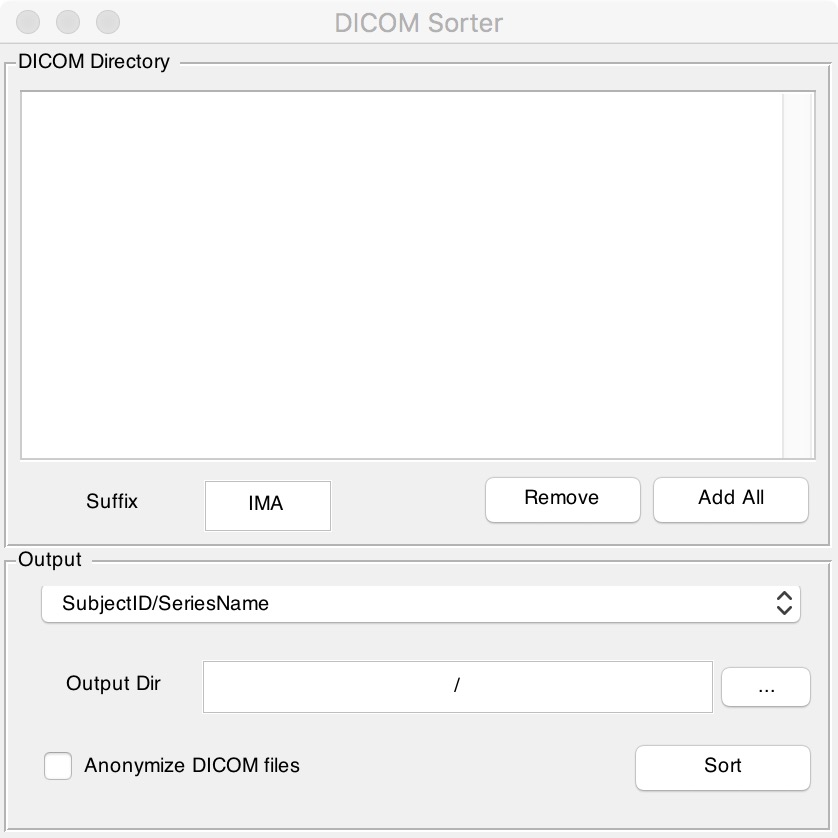
Figure 7. Dicom Sorter
Fill in the suffix of the original file in the "Suffix" box. If there is no suffix, clear this box. Next click "Add All" to find the folder that contains all the original files. Finally enter the output directory you want in "Output Dir" box and click the "Sort" button. It should be noted that if there is no suffix, the original folders should not contain any other types of files except DICOM ones, otherwise you will get an error.
(3) What should I do if “errors” show up?
It's frustrating to see "the red", but you can always log in at http://rfmri.org/DPABIDiscussion, and quickly search by the keyword of the error to see how the previous people solved it. If you do not find the relevant answer, you can also post for help. However, you may need to wait for a while to get the solution. So, before you asking for help, we have a few simple tips for you to take home and worth trying when "the red" shows up.
(1) Run with our Demo Data
Please go to http://rfmri.org/DemoData to download the demo data for processing.
(2) Run with data of one/several subject(s)
You can randomly select one or several subject(s) to run as a test. If your data of these subjects can pass the test, then there is a high probability that the error is caused by the data of some other subjects. At this time, you need to carefully take a look at whether the name of some folder does not correspond strictly, or whether the data of some subject is not copied completely. You can easily infer it from the number of dicom files. Normally, the number of dicom files for each sequence of a same batch of subjects is also the same. You can easily find the problem where the number of dicom files of a subject is inconsistent with others.
(3) Reinstall the software/use another computer
If errors were always reported even though you had tried with data of many subjects, the issue may get a bit tricky. It may be caused by the unfitness of the operating environment. At this time, you can try to uninstall the software, then reinstall it and reboot your computer. Or simply using another computer may resolve this issue. This operating sequence of solutions can be considered: 1) rerun; 2) shutdown and restart matlab; 3) reboot the computer; 4) reinstall DPABI/SPM; 5) reinstall matlab; 6) reinstall windows; 7) try another computer.
DPABI/DPABISurf/DPARSF报错处理
我们注意到,困扰功能磁共振成像初学者最大的问题就是报错(图1)。所有的报错中,电脑环境设置(软件安装)问题占了40%,数据整理的问题占了50%。所以请您务必仔细阅读本指南,因为这将帮助您避免可能遇到的90%的报错!节约您的宝贵时间,让我们有更多时间专注于结果的分析上。

图1. 这种报错(“红字”)你绝不想看到第二遍
1. DPABI/DPABISurf的安装
DPABI/DPABISurf顺利运行的前提包括:
(0)一台性能尚可的windows/mac/linux电脑
处理器intel七代i5或同等水平处理器以上,内存16G以上,预留300G以上的硬盘空间。
(1) MATLAB
因为有很多用户喜欢把SPM或DPABI放在MATLAB的toolbox下,那这样安装路径中不能包括任何空格、汉字或特殊字符!如
“C:\Program files\MATLAB”这样的安装路径就绝对不可以!因为在Program和files之间有一个空格!另外,如果您的操作系统账号名(默认为“Administrator”)包含汉字的话,建议新建一个全英文(没有空格)的账号,将MATLAB安装在该账号下。
(2) SPM
SPM是一个免费的开源软件,请至http://www.fil.ion.ucl.ac.uk/spm/software/download/填写一个简单的个人信息表后下载,建议下载SPM12。切记软件路径中不能包括任何空格、汉字或特殊字符!
(3) FSL (windows用户请忽略)
如果您使用的是mac或者linux系统的电脑,您需要额外安装另外一个磁共振数据处理软件FSL,请至https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FslInstallation按照网页上的步骤完成下载安装。当然,如果您不安装FSL,安装好docker版的DPABISurf也是可以的!
(4)DPABI/DPARSF
请至http://rfmri.org/DPABI下载最新版本的DPABI(包含DPARSF)。
SPM以及DPABI/DPARSF的安装。切记软件路径中不能包括任何空格、汉字或特殊字符!
下载完成后打开MATLAB,点击“Set Path”(图2)。

图2. Set path
首先点击“default”将路径还原,随后点击“Add Folder”来添加SPM路径(SPM千万不要“Add with Subfolders”)。接着点击“Add with Subfolders”(图3),在弹出的窗口中找到存放DPABI的位置,点击“open”添加,最后点击“save”即可关闭此窗口。


图3. add with subfolders
随后在“command window”内输入“dpabi”后回车,当您看到如下图的界面时,就完成DPABI/DPARSF的安装了。



图4. DPABI
2. 数据的整理
DPABI/DPABISurf是一套科学高效的R-fMRI预处理流程,但如果要顺利地让DPABI/DPARSF运作,您必须严格按照DPABI/DPARSF的要求整理好您的数据。提示:您可以用DPABI->Utilities->DPABI Input Preparer来帮助整理数据。在整理数据中,请反复用DPABI->Utilities->Check Data Organization检查数据整理是否合格!几乎所有“索引超出维度”的问题都可以通过Check Data Organization来解决!
(1)建立“工作路径”
在您想要的任意位置建立任意名称的文件夹(但不能包含任何空格、汉字或特殊字符!),这个文件夹就是预处理的“工作路径”,绝大部分预处理操作都在“工作路径”下进行,这是一个很重要的概念,请您牢记。为了示范,我们建立了一个名为“dpabi”的文件夹作为工作路径。

(2)拷贝被试的磁共振数据
接下来我们要按照DPABI/DPARSF的要求将被试的磁共振数据拷贝至工作路径下,以下以三个被试的情况为例:

图中展示了建立好的文件夹的形式,工作路径下应当包含名为“FunRaw”和“T1Raw”的文件夹,每个文件夹下包含三个文件夹:“Sub001”、“Sub002”、“Sub003”,这是三个被试的条件,更多被试的条件请依次类推。
“FunRaw”文件夹下包含的是被试功能像序列的数据,“T1Raw”文件夹下包含的是被试结构像的数据,您需要从原始数据中挑出包含功能像或结构像数据的文件夹,将该文件夹下的数据拷贝至对应的文件夹下。
注意点1: DPABI/DPARSF对工作路径下的数据格式整理要求是非常严格的,“FunRaw”和“T1Raw”这两个文件夹名称必须完全按照规定中的要求,不能有任何变化,否则就会报错。“Sub001”、“Sub002”、“Sub003”这样的被试文件夹虽然比较自由,您可以根据自己的需求修改,但是必须保持“FunRaw”和“T1Raw”两个文件夹下对应的被试文件夹名称完全一致,有任何不同都会导致报错。一个例子是有学员不小心在命名文件夹时在最后多输了一个空格,结果就导致了报错。
注意点2: 这里示范的是当您手中的原始数据是dicom格式的条件下,如果您手中的数据是NIFTI格式的,请将“FunRaw”和“T1Raw”两个文件夹重命名为“FunImg”和“T1Img”。判断dicom格式/NIFTI格式的方法:dicom格式的数据文件数量非常多(成百上千),NIFTI格式的数据文件数量每个被试,每个序列只有一个(4D .nii文件),或数百个(3D .img/.hdr文件)。
注意点3: 一般而言,原始数据是按照序列组织的,每个扫描序列的数据都有对应的文件夹,但有时数据完全是杂乱无章的,这时就需要使用DPABI的“Dicom Sorter”功能了,它可以将杂乱无章的原始数据整理成我们可以理解的形式。
要打开Dicom Sorter,请启动DPABI后依次点击Utilities->DICOM Sorter



“Suffix”栏中填写原始文件的后缀,如果没有后缀就将这一栏清空;点击“Add All”后找到包含所有原始文件的那个文件夹;最后在“Output Dir”中输入您想要输出结果的路径,点击“Sort”键即可,需要注意的是,如果没有后缀的话原始文件夹中不能包含除了DICOM格式之外任何其它类型的文件,否则会报错。
3. 出错了怎么办?
您可以先自行尝试以下几个步骤:
(1)先用我们的演示数据Demo Data跑
请到http://rfmri.org/DemoData下载演示数据进行处理。
(2)单个/若干被试跑
您可以随机抽取出一个被试或者几个被试的数据进行尝试,如果这几个被试可以跑通的话,那就很大几率可以确定问题是出在其它某个被试的身上。这时您就需要仔细检查是不是某个被试的文件夹命名没有严格对应,或者某个被试的数据没有拷贝完全。您可根据dicom文件的数量轻易判断,一般一批被试每个序列dicom文件的数量是一致的,如果您看到有某个被试的dicom文件数目与其他被试的不一致就可以很简单地发现问题所在。
(3)重装软件/换台电脑
如果试了很多个被试都无法跑通,那么问题很有可能比较棘手了,可能是运行环境的问题,这时您可以尝试将软件卸载后重新安装一遍并重启电脑试一下,或者干脆换台电脑,也许就可以解决问题了。可以考虑这样的顺序:1)重新跑;2)重启matlab;3)重启电脑;4)重装DPABI/SPM;5)重装matlab;6)重装windows;7)换电脑。
4. 要提问求助怎么办?
(1) 提问前请仔细学习严老师的免费在线网络课程http://rfmri.org/Course,大多数问题在这里都有答案!认真观看3遍以后再提问。
(2) 请认真阅读严老师的DPABI/DPABISurf/DPARSF报错处理指南https://www.rfmri.org/DPABIErrorHandling,尝试自行排除错误。
(3) 请在在http://rfmri.org搜索相应的报错信息,看看前人是否遇到过相似的错误,严老师是如何回答的,参照解决。
(4) 如果以上方法无法解决你的问题,请在http://rfmri.org提交你的问题。这里面提交的问题肯定都能获得回答的,只是可能会有些滞后。
(5) 建议你选4,不过偶尔也可以在The R-fMRI Journal Club微信群(微信添加rfmriorg2进群)提问。严老师每天早晨统一挑选回答一次。
(6) 如果需要通过参加集中特训解决所有问题,建议参加DPABI/DPABISurf/DAPRSF特训营(http://rfmri.org/DPABICamp)!
祝大家使用DPABI/DPABISurf/DAPRSF愉快!得到好的数据处理结果,做出原创性研究工作,发表高质量的学术论文!



